STARR-seq can be used to test thousands of enhancer candidates in parallel, systematically identify enhancers directly based on their activity across large genomic regions or entire genomes, and to ask fundamental questions of transcriptional regulation and enhancer biology. Below we feature some applications of STARR-seq from others and us (content from Muerdter et al., Genomics 2015).
Genome wide identification of cell-type specific enhancers
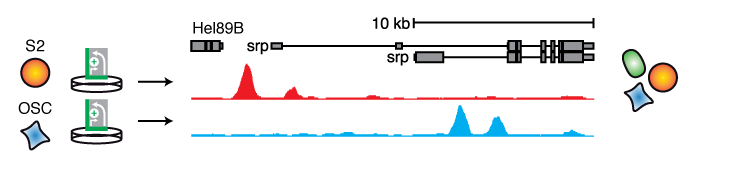
Cell-type specific enhancers drive differential gene expression, hence, identifying such enhancers can be of utmost importance to our understanding of development and differentiation. We generated quantitative enhancer activity maps that describe the enhancer activity landscape of three D. melanogaster cell types of developmentally different origin, one derived from embryos (S2), one from larval brain (BG3), and one from adult ovaries (OSC) by screening the entire fly genome (Arnold 2013; Yanez-Cuna 2014). This revealed thousands of enhancers, exhibiting an activity spectrum ranging from strictly cell type-specific to equally active across cell types and their genomic locations. Importantly, cell type-specific gene expression levels were correlated to the combined activities of the flanking enhancers within each cell type and across cell types, which links differential gene expression to differences in enhancer activity and demonstrates that ectopic assays can accurately assess cell type-specific enhancer activities.
Screening selected genomic loci
STARR-seq can also be applied to libraries of reduced complexity to test selected enhancer candidates (Vanhille 2015; Vockley 2015) or to screen defined genomic regions in an unbiased fashion for active enhancers (Arnold 2013; Zabidi & Arnold 2015; Muerdter & Boryń 2017) or enhancer-activity changes (Rathert 2015).
Pooled enhancer candidate testing

Spicuglia and colleagues captured fragments from sonicated genomic DNA that corresponded to candidate regulatory regions predicted based on chromatin accessibility and TF binding (Vanhille 2015). This allowed them to test 7152 candidate regions in parallel in their murine T-cell model, revealing 2279 weak and 433 strong enhancers, but also demonstrating that many predicted candidates were negative, emphasizing the need for functional validation of such predictions. Reddy and colleagues used STARR-seq to assess the enhancer activities of SNP-containing DNA fragments, thereby determining gene regulatory differences in populations (Vockley 2015).
Screening selected genomic loci
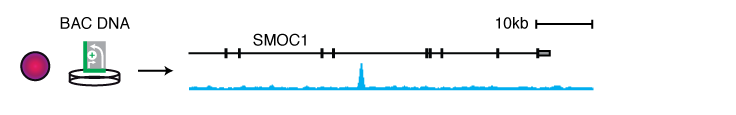
We screened selected genomic regions using libraries derived from randomly fragmented bacterial artificial chromosomes (BACs) to determine enhancer activities (Arnold 2013) and enhancer-activity changes upon cofactor inhibition (Rathert 2015). The latter showed that an enhancer in the Myc locus is strongly activated when the cofactor Brd4 is inhibited by the small molecule inhibitor JQ1. This enhancer contributes to Myc reactivation and survival of JQ1-resistant leukemia cells.
Enhancer activity changes after signaling/treatment - signaling responsive enhancers
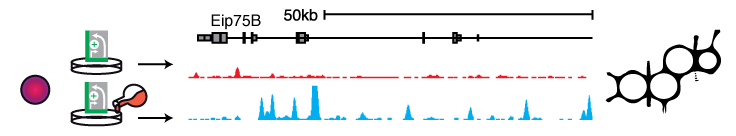
Signaling pathways often cause changes in gene expression levels. Signaling-responsive enhancers direct these changes. STARR-seq allows assessing the underlying differences in enhancer activities by comparing genome-wide enhancer activity maps before and after signal induction. For example, steroid hormones regulate gene expression through their nuclear receptors, which bind to specific DNA motifs and act as transcription factors, thus influencing the activity of the bound enhancer. Using ecdysone signaling in D. melanogaster S2 and OSCs hundreds of hormone dependent, cell type-specific enhancers could be identified and their sequences characterized (Shlyueva 2014). The same experimental setup should also allow for the identification of enhancers downstream of other signaling pathways and enhancer activity changes upon small molecule treatments (e.g. Rathert 2015).
Assessing activity differences of different cis-regulatory genomes – comparative genomics
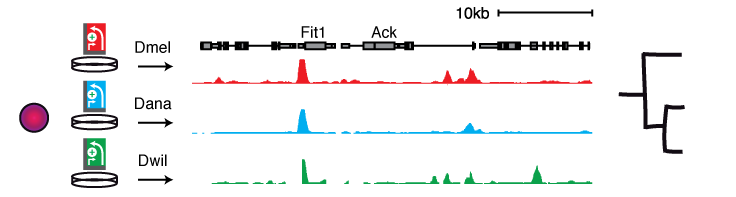
STARR-seq allows screening multiple cis-regulatory genomes in a single cell type, i.e. in the same trans-regulatory environment, enabling powerful comparative analyses of differential enhancer activities that arise from sequence variation.
Screening the genomes of 5 Drosophila species (spanning an evolutionary distance of 30-40 million years) in a single D. melanogaster cell type (S2) revealed that a large portion of D. melanogaster orthologous enhancers is functionally conserved, due to stabilizing turnover of TF motifs. Interestingly, functional enhancers can also be gained within relatively short evolutionary timespans without apparent adaptive selection, yet can be involved in changes of gene expression in vivo (Arnold & Gerlach 2014).
The above approach can be extended to studying sequence variation across different species, within selected populations (see e.g. Vockley 2015), or between DNA from healthy versus disease-tissue (e.g. in cancer) with respect to phenotypic variation or disease.
Functional studies of basic mechanistic questions – enhancer/core-promoter
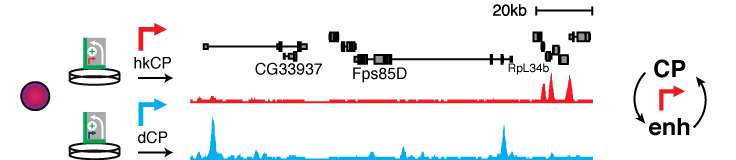
The functional readout and the defined setup of STARR-seq allows asking basic mechanistic questions of enhancer biology. For example, screening the entire fly genome by STARR-seq using different core promoters derived from either ubiquitously active housekeeping genes or from developmentally regulated and cell-type specific genes revealed thousands of enhancers that are specific to either of these two classes of core promoters (Zabidi & Arnold 2015). This suggests the existence of two major transcriptional programs, one for ubiquitous expression of housekeeping genes through ubiquitously active, promoter proximal enhancers and one for the expression of developmental and cell type-specific genes.
This work demonstrates that STARR-seq enables to answer longstanding and fundamental questions of gene regulation and additionally emphasizes the need for powerful functional enhancer assays.
- Content from Muerdter et al., Genomics 2015.