Self-transcribing active regulatory region sequencing (STARR-seq) is a massively parallel reporter assay to identify transcriptional enhancers directly based on their activity in entire genomes and to assess their activity quantitatively (Arnold 2013). Enhancer activity is directly linked to the underlying DNA sequence and measured as presence of the resulting reporter transcripts among cellular RNA by deep sequencing. STARR-seq is complementary to STAP-seq: while STAP-seq measures core-promoter activity using a single defined enhancer, STARR-seq measures enhancer activity using a single defined core-promoter.
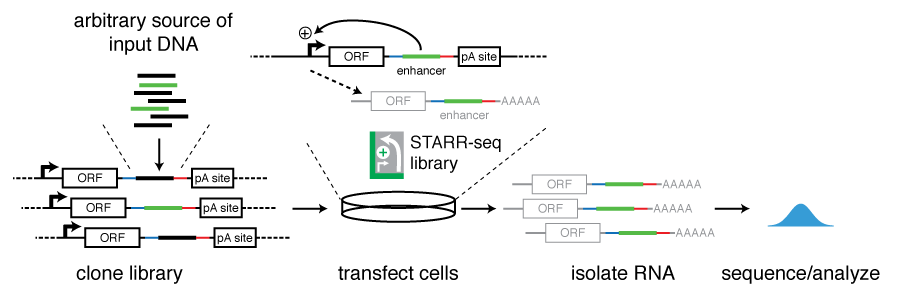
Specifically, DNA fragments are cloned downstream of a core promoter and into the UTR of a reporter gene. Active enhancers will transcribe themselves and become part of the resulting reporter transcripts (see figure). This setup allows the simultaneous testing of millions of DNA sequences in a highly complex reporter library and also ensures that the identified sequences act as bona fide enhancers (rather than for example promoters) as they activate transcription from a remote position.
What are the features of STARR-seq?
As STARR-seq is an ectopic, plasmid-based assay, the measured activity directly reflects the regulatory capacity of enhancer sequences. This measurement is not affected by the location of the candidate sequences within the transcript or their orientation and accurately reflects activity changes after cellular signaling such as hormone treatment highlighting its episomal responsiveness. Due to its episomal nature, STARR-seq is unlikely to suffer from position effects resulting from random genomic integration as has been observed for integrated reporter assays. Furthermore, integration of the reporter constructs in the genome in order to propagate them to daughter cell populations is not necessary due to the short timeframe of STARR-seq, which is in the order of a single cell cycle or less for most cell types.
The activity of candidate sequences tested independently in luciferase assays and STARR-seq is linearly correlated in flies and humans. Thus, STARR-seq reports quantitatively on enhancer activity and constitutes a genome- wide equivalent of luciferase assays. In addition, enhancers identified by STARR-seq are active after random integration into the genome of cell lines, as well as in vivo in transgenic flies after site-specific integration (Arnold 2013). Taken together, STARR-seq draws genome-wide cell type- specific quantitative enhancer activity maps of any cell type that allows efficient delivery of the reporter library.
What are the features of the reporter library?
Candidate DNA fragments can be obtained from arbitrary sources of DNA, including genomic DNA, targeted regions via bacterial artificial chromosomes (BACs), DNA fragments enriched for regions of interest such as open chromatin, TF binding sites, or predicted enhancers as well as synthetic DNA. The size of the candidate DNA fragments can be of a wide range including sizes that fully cover known enhancers. The very large number of input candidate sequences and the wide range of sizes that can be screened place STARR-seq at a unique position among current massively parallel reporter assays and allows unbiased screening of entire genomes.
- Content from Muerdter et al., Genomics 2015.